publications
List of publications in chronological order, including peer-reviewed and preprint papers.
2024
-
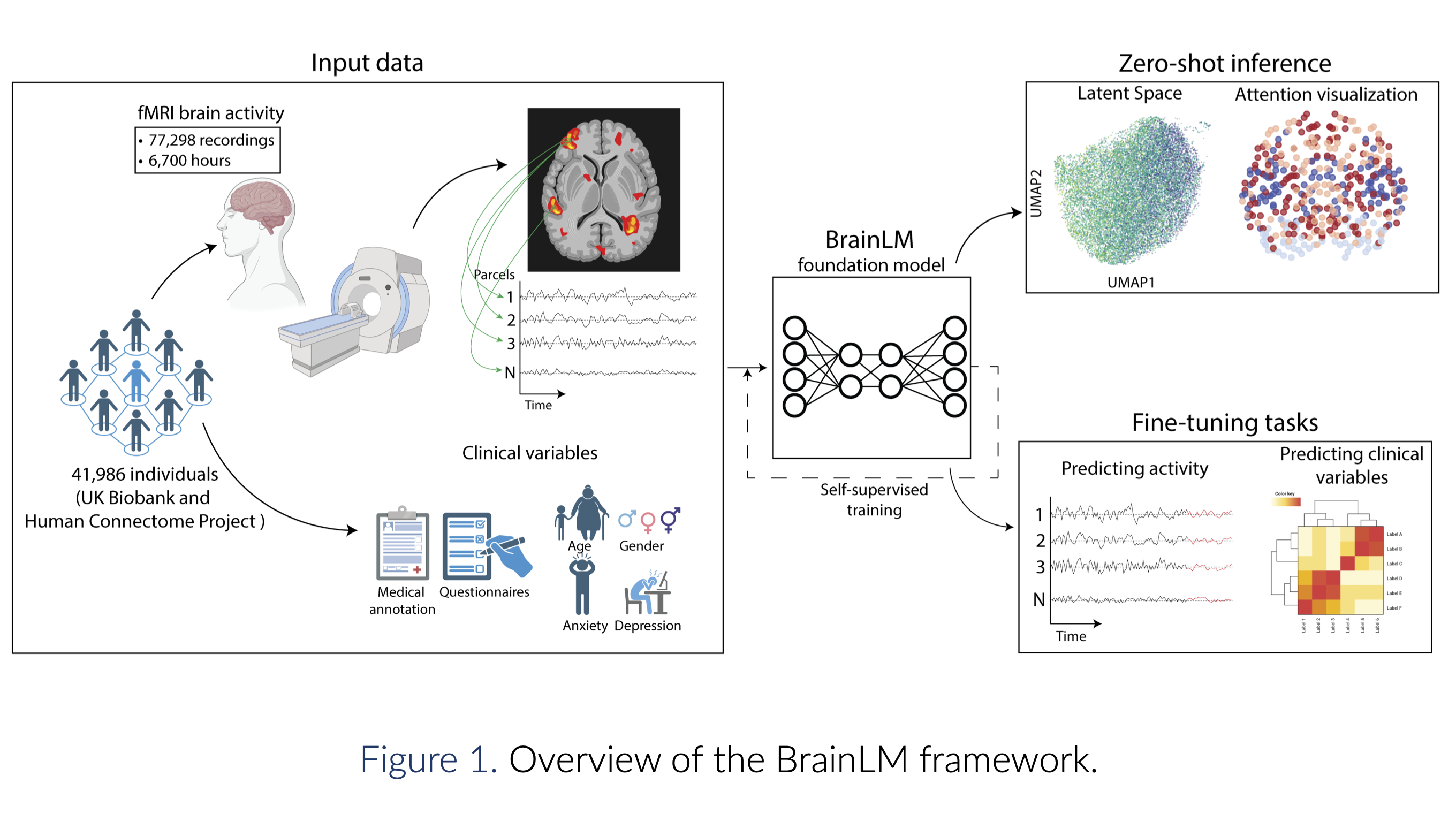 BrainLM: A foundation model for brain activity recordingsJosue Ortega Caro, Antonio Henrique Oliveira Fonseca, Syed A Rizvi, and 8 more authorsIn International Conference on Learning Representations (ICLR) , 2024
BrainLM: A foundation model for brain activity recordingsJosue Ortega Caro, Antonio Henrique Oliveira Fonseca, Syed A Rizvi, and 8 more authorsIn International Conference on Learning Representations (ICLR) , 2024We introduce the Brain Language Model (BrainLM), a foundation model for brain activity dynamics trained on 6,700 hours of fMRI recordings. Utilizing self-supervised masked-prediction training, BrainLM demonstrates proficiency in both fine-tuning and zero-shot inference tasks. Fine-tuning allows for the accurate prediction of clinical variables like age, anxiety, and PTSD as well as forecasting of future brain states. Critically, the model generalizes well to entirely new external cohorts not seen during training. In zero-shot inference mode, BrainLM can identify intrinsic functional networks directly from raw fMRI data without any network-based supervision during training. The model also generates interpretable latent representations that reveal relationships between brain activity patterns and cognitive states. Overall, BrainLM offers a versatile and interpretable framework for elucidating the complex spatiotemporal dynamics of human brain activity. It serves as a powerful "lens" through which massive repositories of fMRI data can be analyzed in new ways, enabling more effective interpretation and utilization at scale. The work demonstrates the potential of foundation models to advance computational neuroscience research.
@inproceedings{caro2023brainlm, title = {BrainLM: A foundation model for brain activity recordings}, author = {Caro, Josue Ortega and de Oliveira Fonseca, Antonio Henrique and Rizvi, Syed A and Rosati, Matteo and Averill, Christopher and Cross, James L and Mittal, Prateek and Zappala, Emanuele and Dhodapkar, Rahul Madhav and Abdallah, Chadi and others}, url = {https://openreview.net/pdf?id=RwI7ZEfR27}, journal = {International Conference on Learning Representations (ICLR)}, booktitle = {International Conference on Learning Representations (ICLR)}, year = {2024}, } -
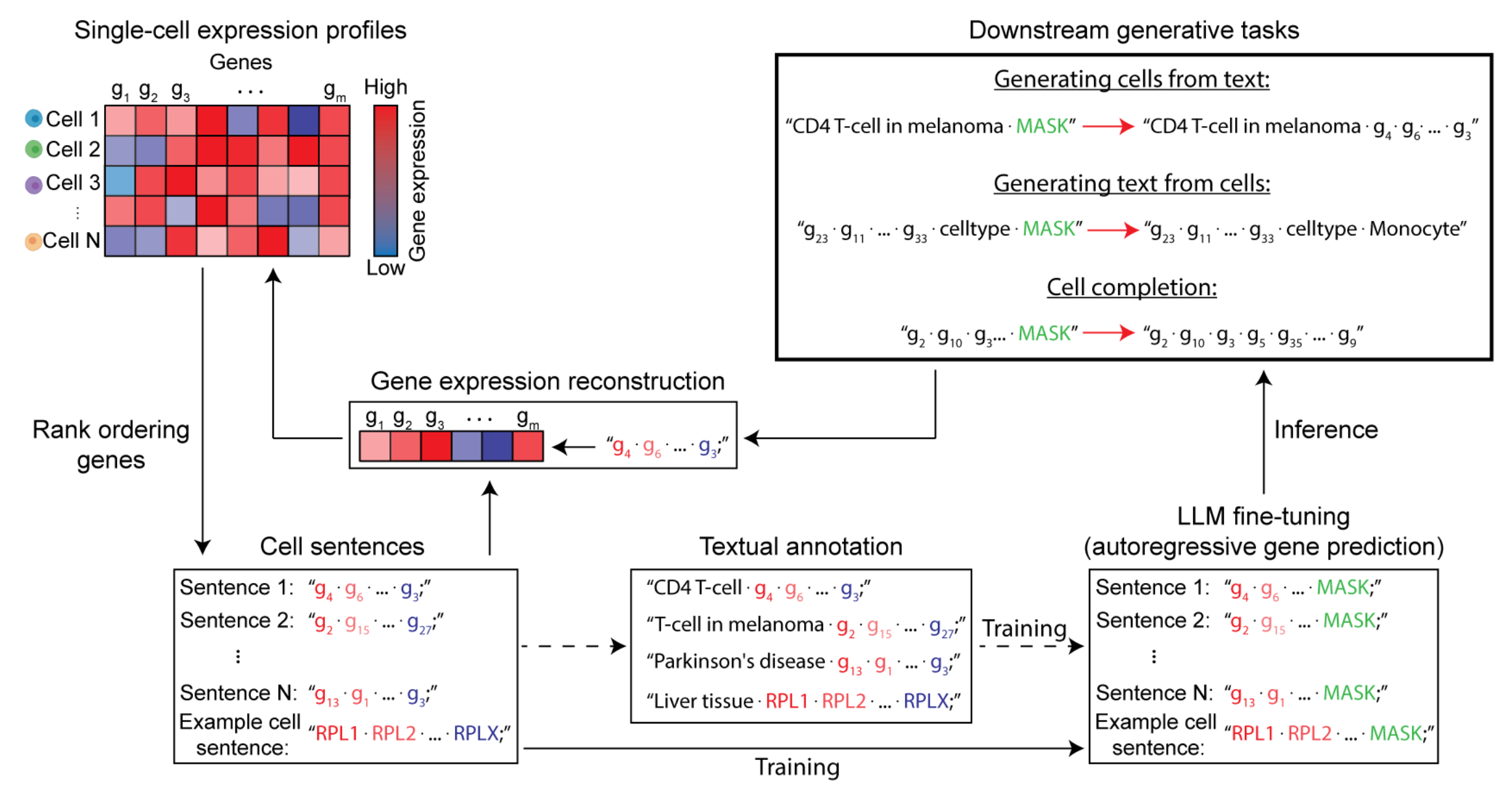 Cell2sentence: Teaching large language models the language of biologyDaniel Levine, Sacha Lévy, Syed Asad Rizvi, and 8 more authorsInternational Conference on Machine Learning (ICML), 2024
Cell2sentence: Teaching large language models the language of biologyDaniel Levine, Sacha Lévy, Syed Asad Rizvi, and 8 more authorsInternational Conference on Machine Learning (ICML), 2024We introduce Cell2Sentence (C2S), a novel method to directly adapt large language models to a biological context, specifically single-cell transcriptomics. By transforming gene expression data into ”cell sentences,” C2S bridges the gap between natural language processing and biology. We demonstrate cell sentences enable the finetuning of language models for diverse tasks in biology, including cell generation, complex celltype annotation, and direct data-driven text generation. Our experiments reveal that GPT-2, when fine-tuned with C2S, can generate biologically valid cells based on cell type inputs, and accurately predict cell types from cell sentences. This illustrates that language models, through C2S finetuning, can acquire a significant understanding of single-cell biology while maintaining robust text generation capabilities. C2S offers a flexible, accessible framework to integrate natural language processing with transcriptomics, utilizing existing models and libraries for a wide range of biological applications.
@article{levine2023cell2sentence, title = {Cell2sentence: Teaching large language models the language of biology}, author = {Levine, Daniel and L{\'e}vy, Sacha and Rizvi, Syed Asad and Pallikkavaliyaveetil, Nazreen and Chen, Xingyu and Zhang, David and Ghadermarzi, Sina and Wu, Ruiming and Zheng, Zihe and Vrkic, Ivan and others}, url = {https://www.biorxiv.org/content/biorxiv/early/2024/02/15/2023.09.11.557287.full.pdf}, journal = {International Conference on Machine Learning (ICML)}, year = {2024}, } -
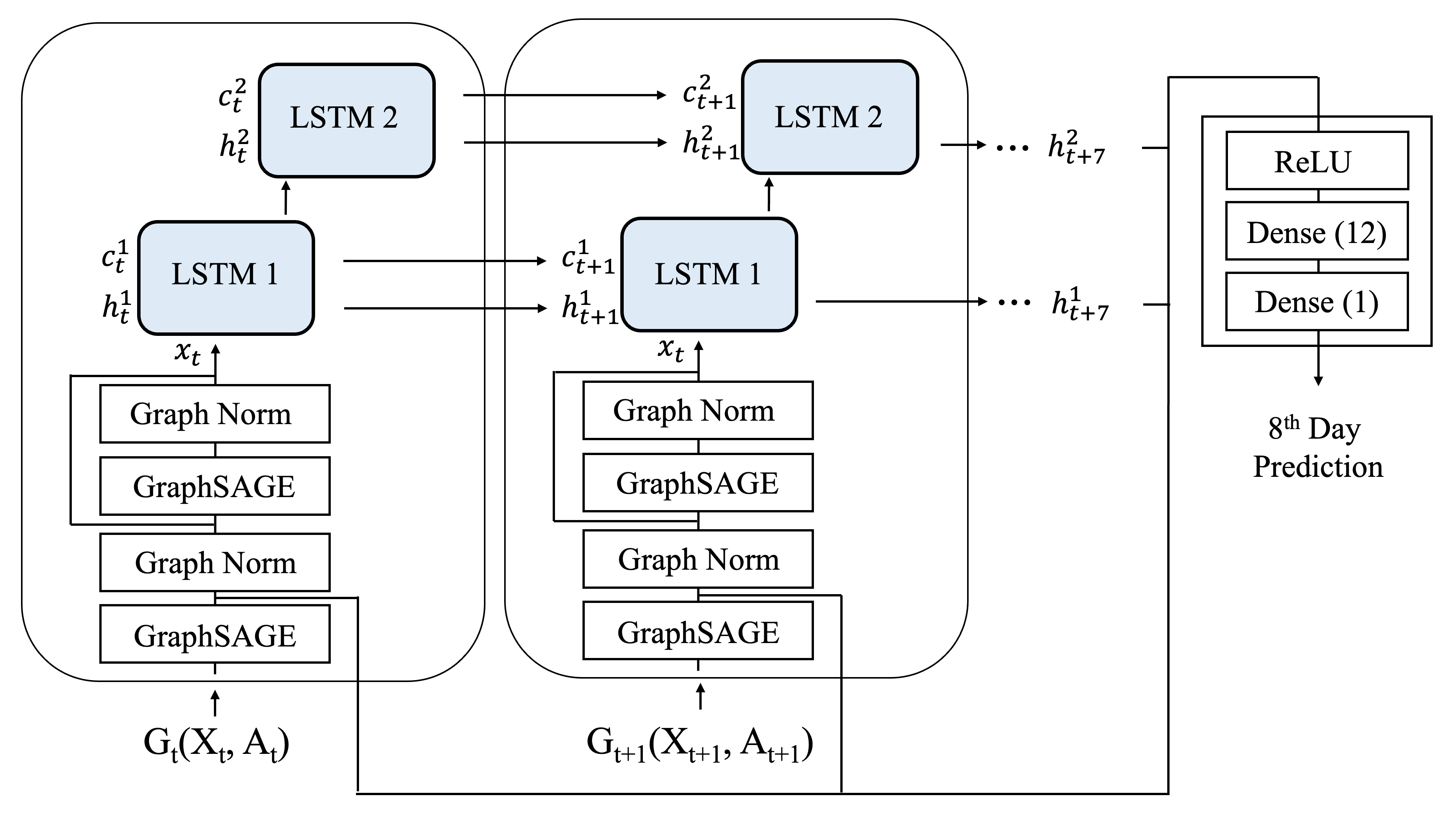 Deep Learning-Derived Optimal Aviation Strategies to Control PandemicsSyed Rizvi, Akash Awasthi, Maria J Peláez, and 4 more authorsNature Scientific Reports, 2024
Deep Learning-Derived Optimal Aviation Strategies to Control PandemicsSyed Rizvi, Akash Awasthi, Maria J Peláez, and 4 more authorsNature Scientific Reports, 2024The COVID-19 pandemic affected countries across the globe, demanding drastic public health policiesto mitigate the spread of infection, which led to economic crises as a collateral damage. In this work,we investigate the impact of human mobility, described via international commercial flights, onCOVID-19 infection dynamics on a global scale. We developed a graph neural network (GNN)-basedframework called Dynamic Weighted GraphSAGE (DWSAGE), which operates over spatiotemporalgraphs and is well-suited for dynamically changing flight information updated daily. This architecture isdesigned to be structurally sensitive, capable of learning the relationships between edge features andnode features. To gain insights into the influence of air traffic on infection spread, we conducted localsensitivity analysis on our model through perturbation experiments. Our analyses identified WesternEurope, the Middle East, and North America as leading regions in fueling the pandemic due to thehigh volume of air traffic originating or transiting through these areas. We used these observationsto propose air traffic reduction strategies that can significantly impact controlling the pandemic withminimal disruption to human mobility. Our work provides a robust deep learning-based tool to studyglobal pandemics and is of key relevance to policymakers for making informed decisions regarding airtraffic restrictions during future outbreaks.
@article{rizvi2022deep, title = {Deep Learning-Derived Optimal Aviation Strategies to Control Pandemics}, author = {Rizvi, Syed and Awasthi, Akash and Pel{\'a}ez, Maria J and Wang, Zhihui and Cristini, Vittorio and Van Nguyen, Hien and Dogra, Prashant}, url = {https://www.nature.com/articles/s41598-024-73639-7.epdf}, journal = {Nature Scientific Reports}, year = {2024}, }
2023
-
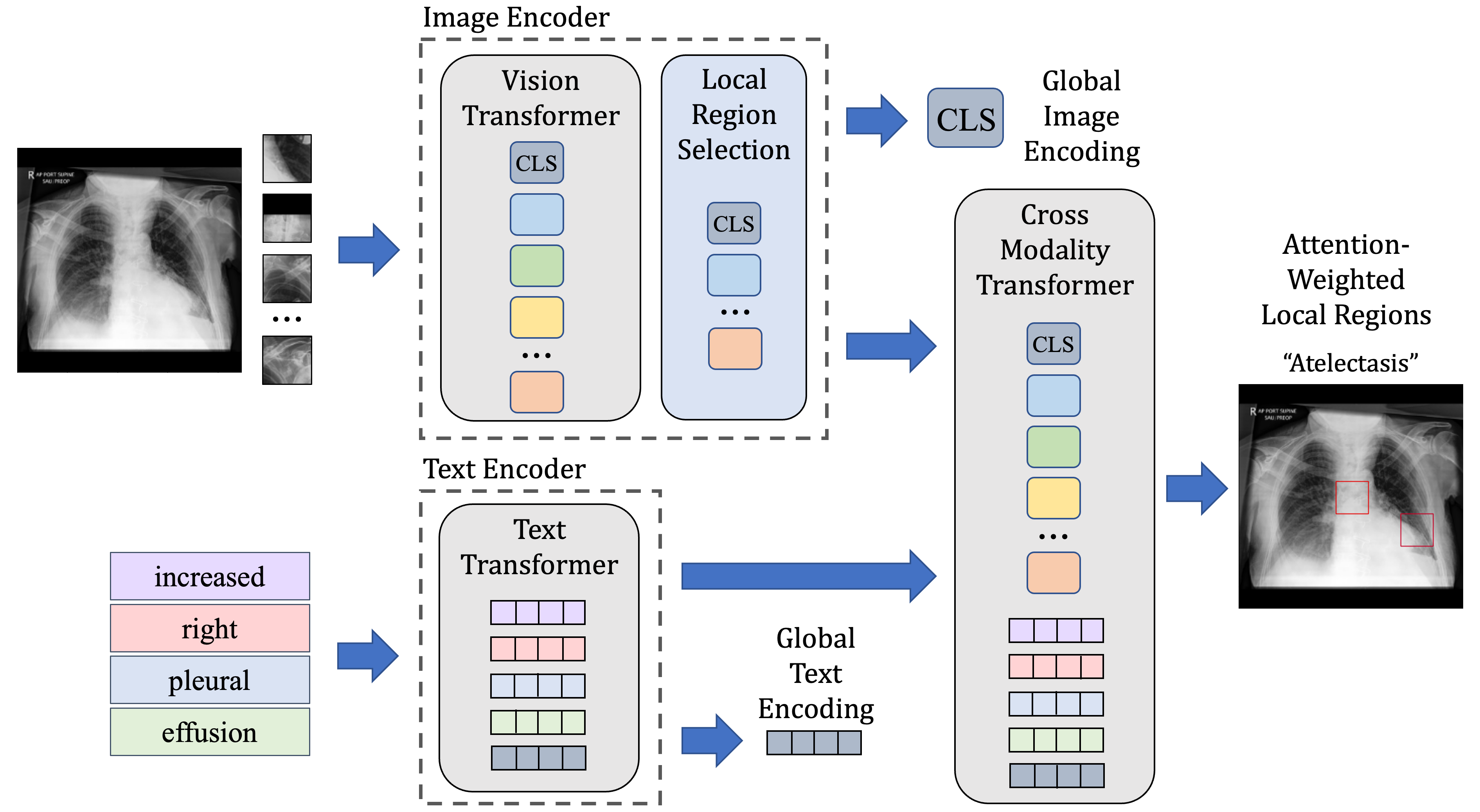 Local contrastive learning for medical image recognitionSyed A Rizvi, Ruixiang Tang, Xiaoqian Jiang, and 2 more authorsIn American Medical Informatics Association (AMIA) Symposium , 2023
Local contrastive learning for medical image recognitionSyed A Rizvi, Ruixiang Tang, Xiaoqian Jiang, and 2 more authorsIn American Medical Informatics Association (AMIA) Symposium , 2023The proliferation of Deep Learning (DL)-based methods for radiographic image analysis has created a great demand for expert-labeled radiology data. Recent self-supervised frameworks have alleviated the need for expert labeling by obtaining supervision from associated radiology reports. These frameworks, however, struggle to distinguish the subtle differences between different pathologies in medical images. Additionally, many of them do not provide interpretation between image regions and text, making it difficult for radiologists to assess model predictions. In this work, we propose Local Region Contrastive Learning (LRCLR), a flexible fine-tuning framework that adds layers for significant image region selection as well as cross-modality interaction. Our results on an external validation set of chest x-rays suggest that LRCLR identifies significant local image regions and provides meaningful interpretation against radiology text while improving zero-shot performance on several chest x-ray medical findings.
@inproceedings{rizvi2023local, title = {Local contrastive learning for medical image recognition}, author = {Rizvi, Syed A and Tang, Ruixiang and Jiang, Xiaoqian and Ma, Xiaotian and Hu, Xia}, url = {https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10785845/}, booktitle = {American Medical Informatics Association (AMIA) Symposium}, volume = {2023}, pages = {1236}, year = {2023}, organization = {American Medical Informatics Association}, } -
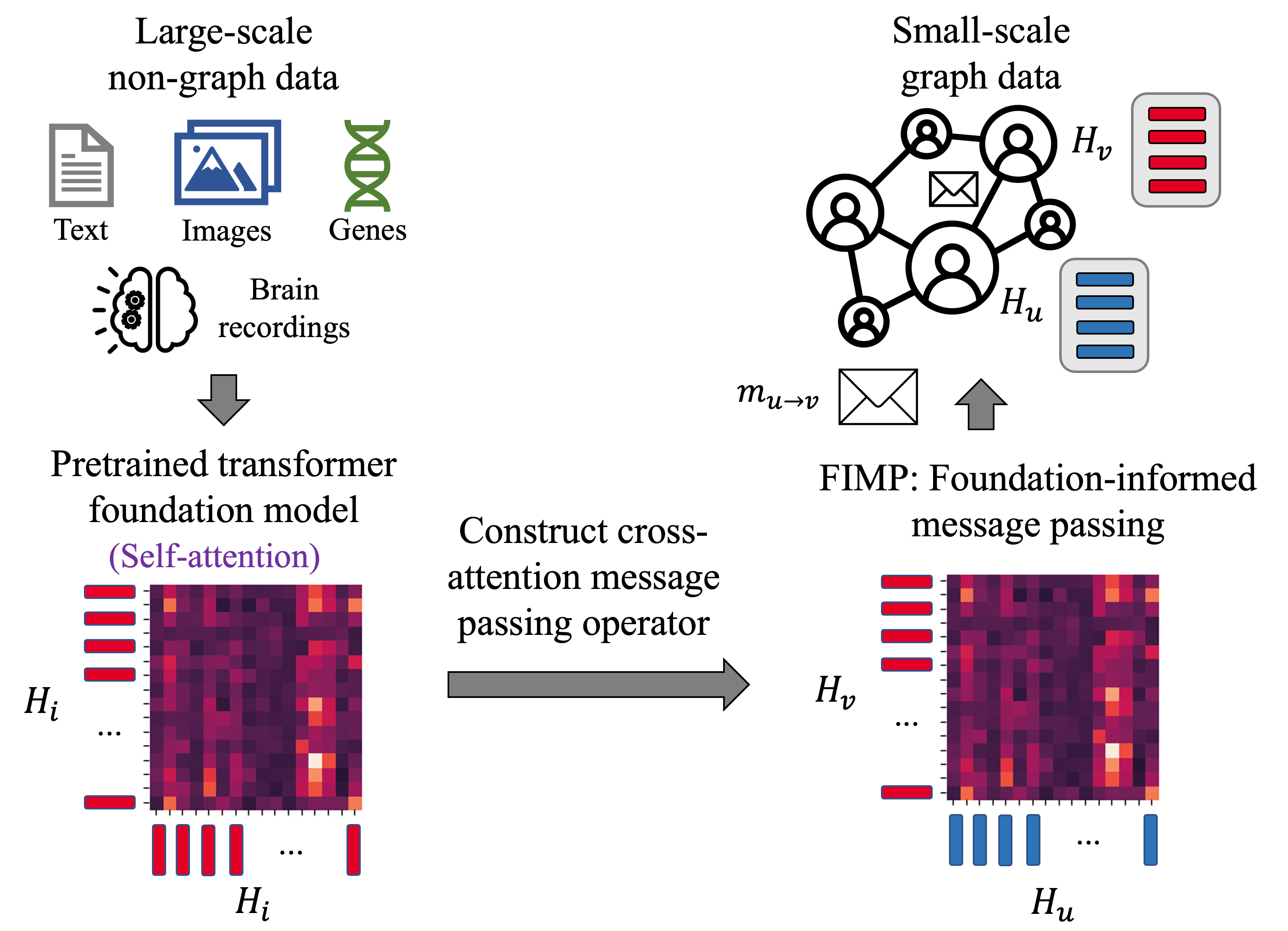 FIMP: Foundation Model-Informed Message Passing for Graph Neural NetworksSyed Asad Rizvi, Nhi Nguyen, Haoran Lyu, and 8 more authorsarXiv preprint arXiv:2210.09475, 2023
FIMP: Foundation Model-Informed Message Passing for Graph Neural NetworksSyed Asad Rizvi, Nhi Nguyen, Haoran Lyu, and 8 more authorsarXiv preprint arXiv:2210.09475, 2023Foundation models have revolutionized the landscape of Deep Learning (DL), serving as a versatile platform which can be adapted to a wide range of downstream tasks. Despite their adaptability, applications of foundation models to downstream graph-based tasks have been limited, and there remains no convenient way to leverage large-scale non-graph pretrained models in graph-structured settings. In this work, we present a new framework which we term Foundation-Informed Message Passing (FIMP) to bridge the fields of foundational models and GNNs through a simple concept: constructing message-passing operators from pretrained foundation model weights. We show that this approach results in improved performance for graph-based tasks in a number of data domains, allowing graph neural networks to leverage the knowledge of foundation models.
@article{rizvi2022fimp, title = {FIMP: Foundation Model-Informed Message Passing for Graph Neural Networks}, author = {Rizvi, Syed Asad and Nguyen, Nhi and Lyu, Haoran and Christensen, Benjamin and Caro, Josue Ortega and Fonseca, Antonio HO and Zappala, Emanuele and Bagherian, Maryam and Averill, Christopher and Abdallah, Chadi G and others}, url = {https://arxiv.org/pdf/2210.09475}, journal = {arXiv preprint arXiv:2210.09475}, year = {2023}, }
2022
-
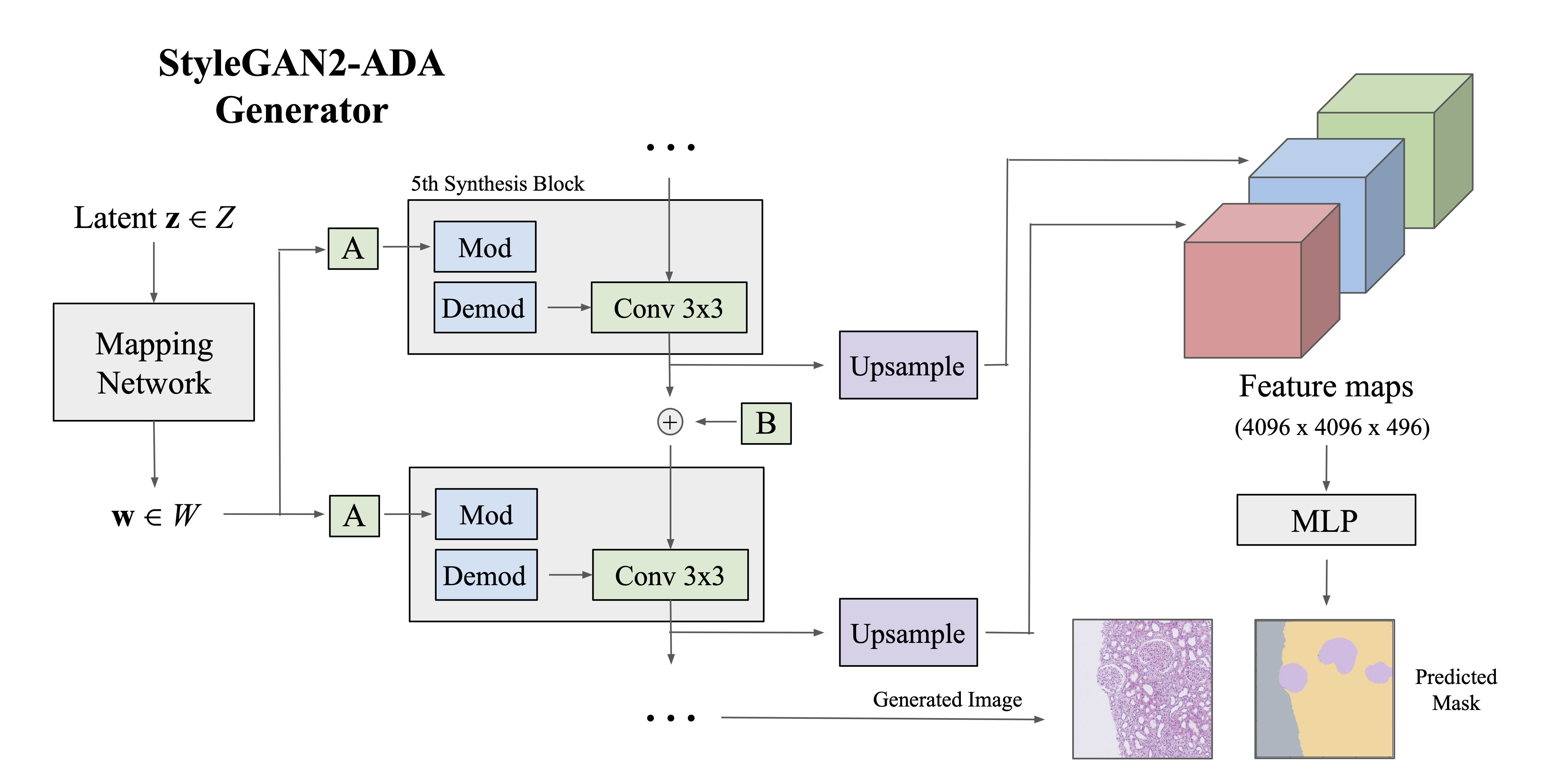 Histopathology DatasetGAN: Synthesizing Large-Resolution Histopathology DatasetsS Rizvi, P Cicalese, S Seshan, and 3 more authorsIEEE Signal Processing in Medicine and Biology (SPMB), 2022
Histopathology DatasetGAN: Synthesizing Large-Resolution Histopathology DatasetsS Rizvi, P Cicalese, S Seshan, and 3 more authorsIEEE Signal Processing in Medicine and Biology (SPMB), 2022Deep learning-based methods have powered recent advancements in medical image segmentation, accelerating the field past previous statistical and Machine Learning-based methods [1]. This, however, has simultaneously created a need for large quantities of labeled data, which is difficult in domains such as medical imaging where labeling is expensive and requires expert knowledge. Semi-supervised learning (SSL) addresses these limitations by augmenting labeled data with large quantities of more widely available unlabeled data. Existing semi-supervised frameworks based on pseudo-labeling [2] or contrastive methods [3], however, struggle to scale to the high resolution of medical image datasets. In this work, we propose the Histopathology DatasetGAN (HDGAN) framework, an extension of the DatasetGAN framework for image generation and segmentation that scales well to large-resolution histopathology images. We make several adaptations on the original framework, including updating the generative backbone, selectively extracting latent features from the generator, and switching to memory-mapped arrays. These changes reduce the memory consumption of the framework, improving its applicability to medical imaging domains.
@article{rizvihistopathology, title = {Histopathology DatasetGAN: Synthesizing Large-Resolution Histopathology Datasets}, author = {Rizvi, S and Cicalese, P and Seshan, S and Sciascia, S and Becker, J and Nguyen, H}, url = {https://isip.piconepress.com/conferences/ieee_spmb/2022/papers/p01_16.pdf}, journal = {IEEE Signal Processing in Medicine and Biology (SPMB)}, year = {2022}, }
2021
-
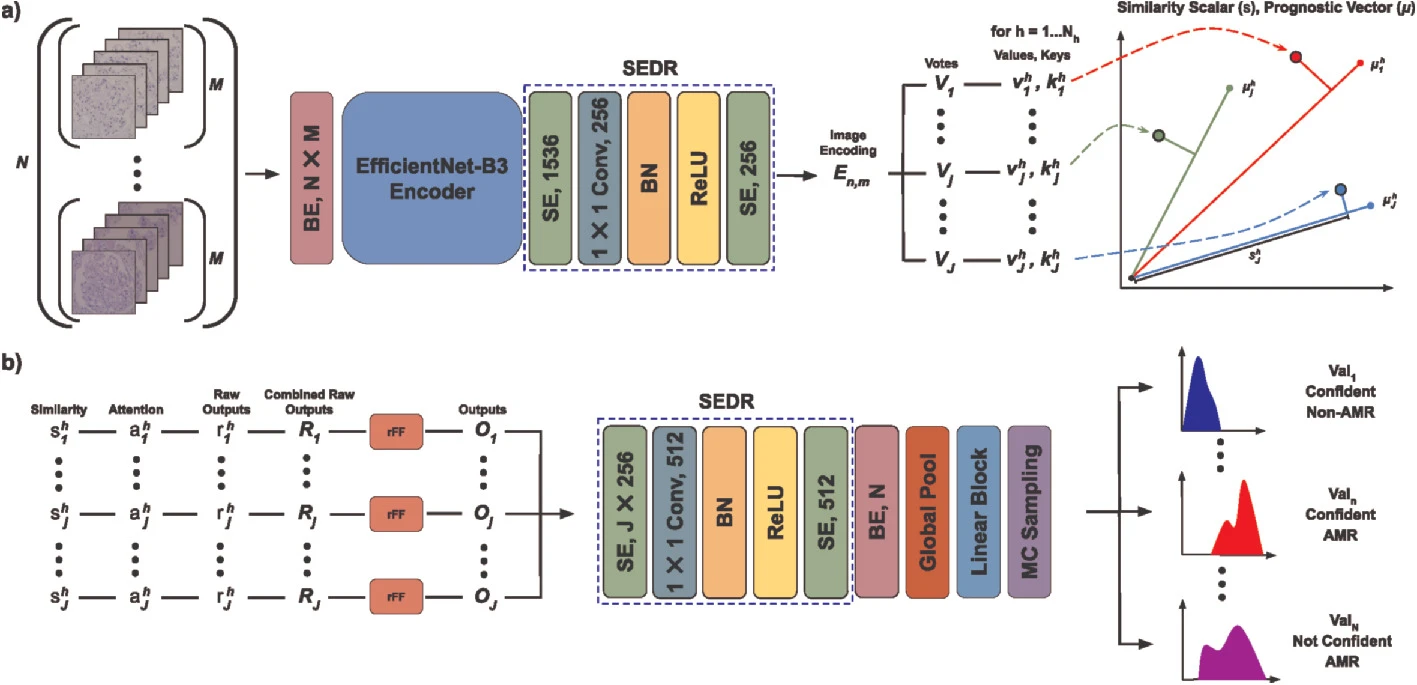 MorphSet: Improving Renal Histopathology Case Assessment Through Learned Prognostic VectorsPietro Antonio Cicalese, Syed Asad Rizvi, Victor Wang, and 8 more authorsIn Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, September 27–October 1, 2021, Proceedings, Part VIII 24 , 2021
MorphSet: Improving Renal Histopathology Case Assessment Through Learned Prognostic VectorsPietro Antonio Cicalese, Syed Asad Rizvi, Victor Wang, and 8 more authorsIn Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, September 27–October 1, 2021, Proceedings, Part VIII 24 , 2021Computer Aided Diagnosis (CAD) systems for renal histopathology applications aim to understand and replicate nephropathologists’ assessments of individual morphological compartments (e.g. glomeruli) to render case-level histological diagnoses. Deep neural networks (DNNs) hold great promise in addressing the poor intra- and interobserver agreement between pathologists. This being said, the generalization ability of DNNs heavily depends on the quality and quantity of training labels. Current “consensus” labeling strategies require multiple pathologists to evaluate every compartment unit over thousands of crops, resulting in enormous annotative costs. Additionally, these techniques fail to address the underlying reproducibility issues we observe across various diagnostic feature assessment tasks. To address both of these limitations, we introduce MorphSet, an end-to-end architecture inspired by Set Transformers which maps the combined encoded representations of Monte Carlo (MC) sampled glomerular compartment crops to produce Whole Slide Image (WSI) predictions on a case basis without the need for expensive fine-grained morphological feature labels. To evaluate performance, we use a kidney transplant Antibody Mediated Rejection (AMR) dataset, and show that we are able to achieve 98.9% case level accuracy, outperforming the consensus label baseline. Finally, we generate a visualization of prediction confidence derived from our MC evaluation experiments, which provides physicians with valuable feedback.
@inproceedings{cicalese2021morphset, title = {MorphSet: Improving Renal Histopathology Case Assessment Through Learned Prognostic Vectors}, author = {Cicalese, Pietro Antonio and Rizvi, Syed Asad and Wang, Victor and Patibandla, Sai and Yuan, Pengyu and Zare, Samira and Moos, Katharina and Batal, Ibrahim and Clahsen-van Groningen, Marian and Roufosse, Candice and others}, url = {https://link.springer.com/chapter/10.1007/978-3-030-87237-3_31}, booktitle = {Medical Image Computing and Computer Assisted Intervention--MICCAI 2021: 24th International Conference, Strasbourg, France, September 27--October 1, 2021, Proceedings, Part VIII 24}, pages = {319--328}, year = {2021}, organization = {Springer}, }